useR to programmeR
Functions 2
Learning objectives
In this session, we will discuss:
- embracing
{{}}for<data-masking>functions - tidyverse style and design of functions
- the joys of side-effects
For coding, we will use r-programming-exercises:
-
R/functions-02-01-embrace.R, etc. - with each new file, restart R.
Plot functions: Motivation
Sometimes, you want to generalize a certain type of plot - let’s say a histogram:
diamonds |>
ggplot(aes(x = carat)) +
geom_histogram(binwidth = 0.1)
diamonds |>
ggplot(aes(x = carat)) +
geom_histogram(binwidth = 0.05)“I want choose only the data, variable, and bin-width”
Function
aes() is a data-masking function; you can embrace 🤗
- pass “bare-name” variables for data-frames
- look for
<data-masking>in help
histogram <- function(df, var, binwidth = NULL) {
df |>
ggplot(aes(x = {{ var }})) +
geom_histogram(binwidth = binwidth)
}
histogram(diamonds, carat, 0.1) Our turn
Complete this function yourself:
histogram <- function(df, var, binwidth = NULL) {
df |>
ggplot(aes()) +
geom_histogram(binwidth = binwidth)
}Try with other
dfandvar, e.g.starwars,mtcars.-
Using this function as is, how can you:
add
theme_minimal()?fill the bars with
"steelblue"?
Our turn (solution)
Adding a theme: histogram() returns a ggplot object, so you can add a theme in the “usual” way:
histogram(starwars, height) + theme_minimal()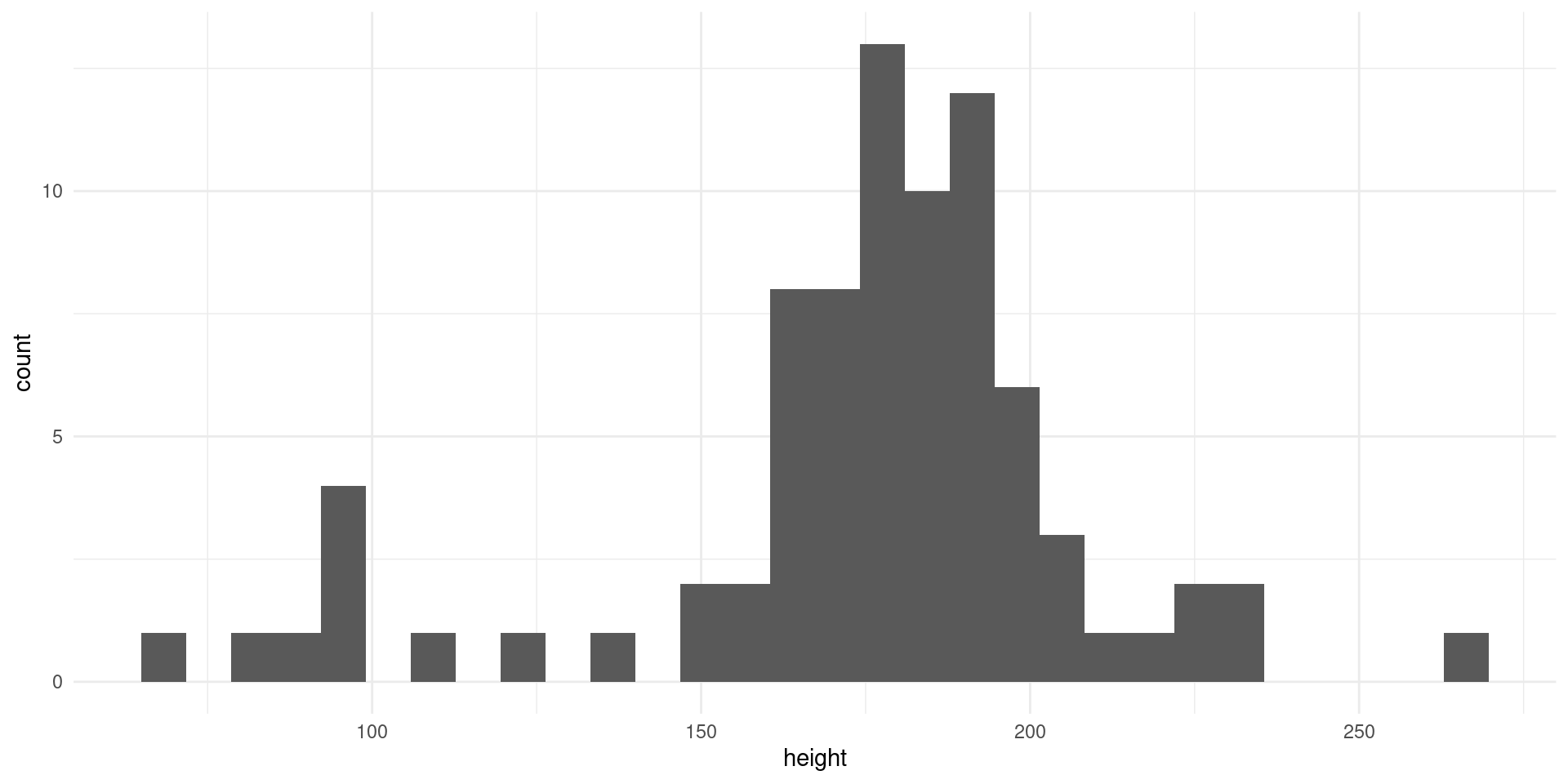
Our turn (solution)
As is, there is no easy way to specify "steelblue".
However, you can build an escape hatch.
... are called “dot-dot-dot” or “dots”.
# `...` passed to `geom_histogram()`
histogram <- function(df, var, ..., binwidth = NULL) {
df |>
ggplot(aes(x = {{var}})) +
geom_histogram(binwidth = binwidth, ...)
}Passes unspecified arguments from your function to another (tell your users where).
Tidyverse Design Guide has more details.
Our turn (continued)
Incorporate dot-dot-dot into histogram():
histogram <- function(df, var, ..., binwidth = NULL) {
df |>
ggplot(aes(x = {{var}})) +
geom_histogram(binwidth = binwidth, ...)
}Try, e.g.:
histogram(starwars, height, binwidth = 5, fill = "steelblue")Tradeoffs
You write a function to make some things easier.
The cost is that some things become more difficult.
This is unavoidable, the best you can do is be deliberate about what you make easier and more difficult.
Labelling
How to build a string, using variable-names and values?
rlang::englue() was built for this purpose:
- embrace 🤗 variable-names:
{{}} - glue values:
{}
temp <- function(varname, value) {
rlang::englue("You chose varname: {{ varname }} and value: {value}")
}
temp(val, 0.4)[1] "You chose varname: val and value: 0.4"Your turn
Adapt histogram() to include a title that describes:
- which
varis binned, and thebinwidth
histogram <- function(df, var, ..., binwidth = NULL) {
df |>
ggplot(aes(x = {{ var }})) +
geom_histogram(binwidth = binwidth, ...) +
labs(
title = rlang::englue("")
)
}Try:
histogram(starwars, height, binwidth = 5)
histogram(starwars, height) # "extra credit"Your turn (solution)
histogram <- function(df, var, ..., binwidth = NULL) {
df |>
ggplot(aes(x = {{ var }})) +
geom_histogram(binwidth = binwidth, ...) +
labs(
title = rlang::englue(
"Histogram of {{ var }}, with binwidth {binwidth %||% 'default'}"
)
)
}
histogram(starwars, height, binwidth = 5)Mixing in other tidyverse functions
Your function can also include pre-processing of data.
Mixing in other tidyverse functions
Your function can also include pre-processing of data.
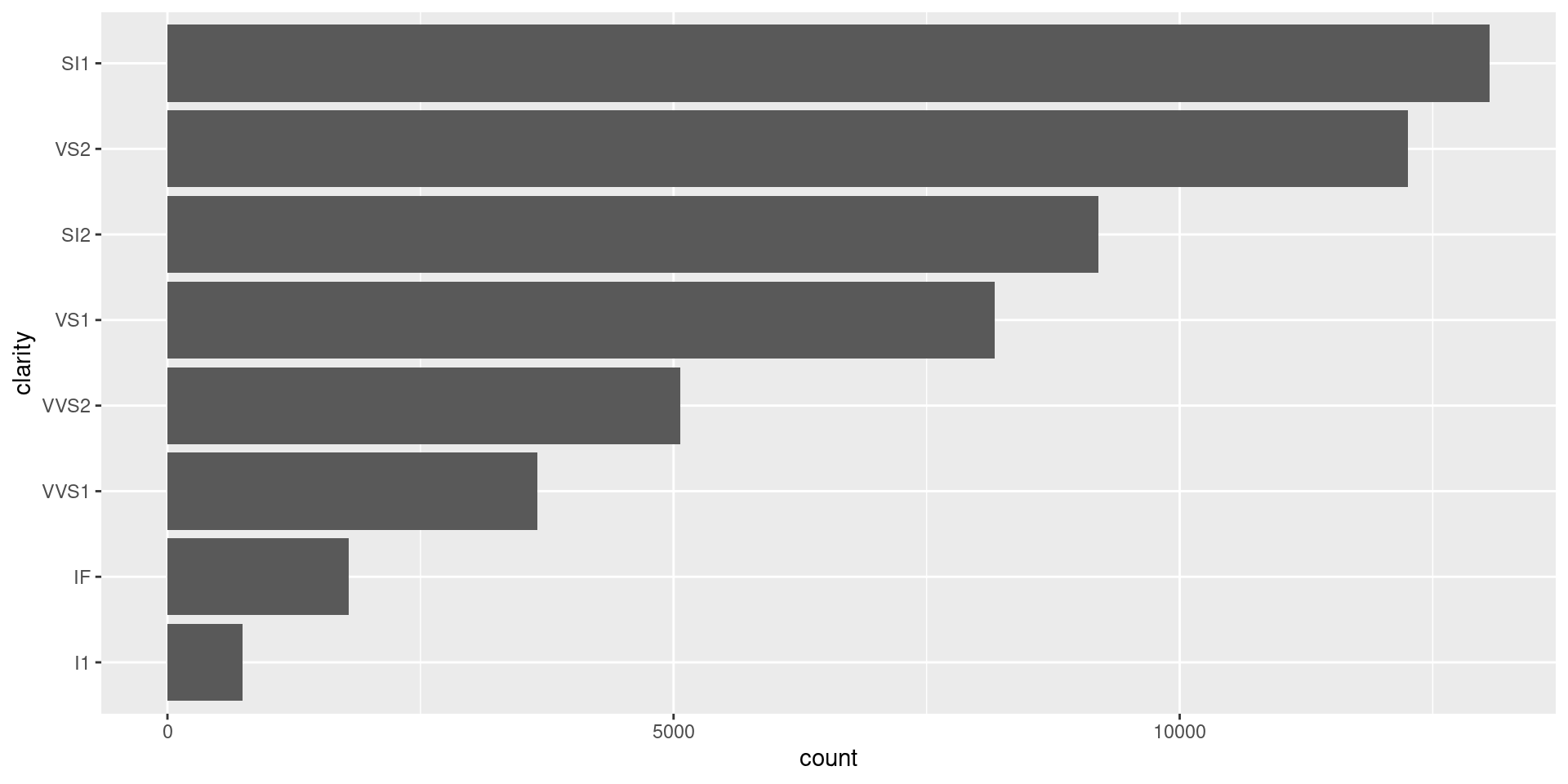
sorted_bars <- function(df, var) {
df |>
mutate({{ var }} := {{ var }} |> fct_infreq() |> fct_rev()) |>
ggplot(aes(y = {{ var }})) +
geom_bar()
}If using
{{ }}to specify a new column, use:=, not=.fct_infreq()reorders by decreasing frequency.fct_rev()reverses order, as y-axis starts at bottom.Our turn: let’s try it
Summary (so far)
-
use embracing
{{}}to interpolate bare column-names- function receiving the “embracing” has to be aware
- look for
<data-masking>in the help - use
rlang::englue()to interpolate variables{{}}and values{}
-
...is a useful “escape hatch” in function design:- put after required args, and before details
- tell your users where the dots are going
Design and style
Restart R, open functions-02-02-style.R
Use descriptive name, usually starts with a verb, unless it returns a well-known noun.
Order of arguments
required: arguments without default values
dots: can be passed on functions that your function calls
optional: arguments with default values
Tidyverse Design:
Order of arguments
Our histogram function:
histogram <- function(df, var, ..., binwidth = NULL) {
df |>
ggplot(aes(x = {{var}})) +
geom_histogram(binwidth = binwidth, ...)
}required:
df,vardots:
...optional:
binwidth
Order of arguments
histogram <- function(df, var, ..., binwidth = NULL) {
df |>
ggplot(aes(x = {{ var }})) +
geom_histogram(binwidth = binwidth, ...)
}Why optional after dots?
user must name optional arguments, in this case
binwidth.makes code easier to read when optional arguments used.
more reasoning given in the Tidyverse design guide.
Namespacing functions
When we write filter(), do we mean…
Three ways to sort this out:
library("conflicted"), suitable for R scriptspackage::function(), used in package functions#' @importFrom, also used (sparingly) in packages
📦 conflicted
conflicted lets you know when you use a function that exists two-or-more packages that you’ve loaded.
To avoid conflicts, declare a preference:
# put in a conspicuous place, near the top of your script
conflicts_prefer(dplyr::filter)Your turn
In functions-02-02-style.R:
library("tidyverse")
mtcars |> filter(cyl == 6)- run it as-is
- add
library("conflicted"), run again - add a
conflicts_prefer()directive
package::function()
This is the usual way when writing a function for a package:
histogram <- function(df, var, ..., binwidth = NULL) {
df |>
ggplot2::ggplot(ggplot2::aes(x = {{ var }})) +
ggplot2::geom_histogram(binwidth = binwidth, ...)
}- makes it very clear where you are getting the function from
- can be verbose, especially if calling an external function often
There is a balance to be struck.
#' @importFrom
When you have a lot of calls to a given external function
Put this in {packagename}-package.R:
#' @importFrom ggplot2 ggplot aes geom_histogram
NULLAlternatively, from the R command prompt:
usethis::use_import_from("ggplot2", c("ggplot", "aes", "geom_histogram"))#' @importFrom
histogram <- function(df, var, ..., binwidth = NULL) {
df |>
ggplot(aes(x = {{ var }})) +
geom_histogram(binwidth = binwidth, ...)
}Makes your code less verbose, but also less transparent
To mitigate:
- put all your
@importFromin one conspicuous file:{packagename}-package.R - use judiciously
Design and style references
Also, look at tidyverse code at GitHub (my favorite is {usethis})
Side effects
Restart R, open functions-02-03-side-effects.R
Uses side effects:
Depends on something other than inputs, e.g.
read.csv()Or, makes a change in the environment, e.g.
print()
Pure function
add <- function(x, y) {
x + y
}The return value depends only on the inputs.
Easier to test.
Uses side effects
Side-effects can slow down your function:
- it can be costly to read/write to disk, print to the screen.
Depending on side effects can introduce uncertainty:
- are you certain of what
file.csvcontains?
Side effects aren’t necessarily bad, but you need to take them into account:
- need to take care when testing.
Your turn
Discuss with your neighbor, are these function-calls are pure, or do they use side effects?
In functions-02-03-side-effects.R:
Our turn: checking locale
Can be useful to consult devtools::session_info():
# using `info = "platform"` to fit output on screen
devtools::session_info(info = "platform")─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.3.1 (2023-06-16)
os Ubuntu 22.04.3 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate C.UTF-8
ctype C.UTF-8
tz UTC
date 2023-09-18
pandoc 3.1.1 @ /opt/quarto/bin/tools/ (via rmarkdown)
──────────────────────────────────────────────────────────────────────────────Manage side effects using 📦 withr
Side effects can include:
- modifying environment:
Sys.setenv() - modifying options:
options() - setting random seed:
set.seed() - setting working directory:
setwd() - creating and writing to a temporary file
{withr} makes it a lot easier to “leave no footprints”.
Our turn: modifying locale
sort() uses locale (environment) for string-sorting rules
(temp <- Sys.getlocale("LC_COLLATE"))
## [1] "C.UTF-8"
sort(c("apple", "Banana", "candle"))
## [1] "apple" "Banana" "candle"Sys.setlocale("LC_COLLATE", "C")
## [1] "C"
sort(c("apple", "Banana", "candle"))
## [1] "Banana" "apple" "candle"
Sys.setlocale("LC_COLLATE", temp)
## [1] "C.UTF-8"Our turn: setting within call
To temporarily set locale:
withr::with_locale(
new = list(LC_COLLATE = "C"),
sort(c("apple", "Banana", "candle"))
)
## [1] "Banana" "apple" "candle"
Sys.getlocale("LC_COLLATE")
## [1] "C.UTF-8"Our turn: setting only within scope
c_sort <- function(...) {
# set only within function block
withr::local_locale(list(LC_COLLATE = "C"))
sort(...)
}
c_sort(c("apple", "Banana", "candle"))
## [1] "Banana" "apple" "candle"
Sys.getlocale("LC_COLLATE")
## [1] "C.UTF-8"Within curly brackets applies to function blocks, it also applies to {testthat} blocks.
But what about dplyr?
?dplyr::arrange()-
arrange()uses the"C"locale by default
Your turn
library("testthat")
test_that("mtcars has expected columns", {
expect_type(mtcars$cy, "double")
})Test passed 🌈This passes, but R is doing partial matching on the $.
Modify test_that() block to warn on partial matching.
You can get the current setting using:
getOption("warnPartialMatchDollar")[1] FALSEHint: use withr::local_option().
Your turn (solution)
test_that("mtcars has expected columns", {
withr::local_options(list(warnPartialMatchDollar = TRUE))
expect_type(mtcars$cy, "double")
})── Warning ('<text>:5:3'): mtcars has expected columns ─────────────────────────
partial match of 'cy' to 'cyl'
Backtrace:
1. testthat::expect_type(mtcars$cy, "double")
2. testthat::quasi_label(enquo(object), arg = "object")
3. rlang::eval_bare(expr, quo_get_env(quo))And yet…
getOption("warnPartialMatchDollar")[1] FALSESummary
You can use tidy evaluation in {ggplot2} to specify aesthetics, add labels, and include {dplyr} preprocessing:
- embracing
{{}}for<data-masking>functions - be aware of
<tidy-select>functions, work differently
Using tidyverse style and design can make things easier for you, your users, and future you.
Be mindful of side effects, use {withr} to manage global state.