usethis::use_course("posit-conf-2023/programming-r-exercises")useR to programmeR
👋 & Functions 1
WiFi: Posit Conf 2023
Password: conf2023
👋 Welcome
Introductions
This is a two-day, hands-on workshop for those who have embraced the tidyverse and want to improve their R programming skills and, especially, reduce the amount of duplication in their code.
- do you have experience equivalent to an introductory data science course using tidyverse?
- are you comfortable with the Whole game chapter of R for Data Science (2nd Edition) by by Hadley Wickham, Mine Çetinkaya-Rundel, and Garrett Grolemund.
Material
- pos.it/programming-r-conf-2023
The team
Standing on the shoulders of
R for Data Science (2e) Wickham, Çetinkaya-Rundel, and Grolemund (2023)
The tidyverse style guide Wickham (n.d.)
Programming with dplyr vignette Wickham et al. (2022)
WiFi
- Network: Posit Conf 2023
- Password: conf2023
Introductions
To each other! With help from Yorkshire!

Ingredients: Sugar, Glucose syrup, Cocoa mass, Vegetable fats (Palm, Rapeseed, Sunflower, Coconut,Mango kernel/ Sal/ Shea), Sweetened condensed skimmed milk (Skimmed milk, Sugar), Cocoa butter, Dried whole milk, Glucose-fructose syrup, Coconut, Lactose and proteins from whey (from Milk), Whey powder (from Milk), Hazelnuts, Skimmed milk powder, Butter (from Milk), Emulsifiers (Sunflower lecithin, E471), Flavourings, Butterfat (from Milk), Fat-reduced cocoa powder, Salt, Lactic acid.

Ingredients: Sugar, Semi-Sweet Chocolate (Sugar, Chocolate, Cocoa Butter, Milkfat, Soy and Sunflower Lecithin, Natural Vanilla Flavor), Glucose Syrup, Peppermint Oil, Citric Acid, Invertase.

Ingredients: Glucose Syrup, Sugar, Starch, Acid: Citric Acid, Flavouring, Fruit and Plant Concentrates: Aronia, Blackcurrant, Elderberry, Grape, Lemon, Orange, Safflower Spirulina, Caramelised Sugar Syrup, Glazing Agents: Beeswax, Carnauba Wax, Elderberry Extract.
Code of Conduct
Code of Conduct and COVID policies. Please Review
- 💙 Treat everyone with respect
- 🧡 Everyone should feel welcome and safe
Reporting:
- 🗣️ any posit::conf staff member (t-shirt) or Info desk
- 📧
conf@posit.com - ☎️ 844-448-1212
Housekeeping
- There are gender-neutral bathrooms located are among the Grand Suite Bathrooms.
- Meditation/prayer rooms: Grand Suite 2A and Grand Suite 2B. Open Sunday - Tuesday 07:30 - 19:00, Wednesday 08:00 - 18:00
- Lactation room: in Grand Suite 1. Open Sunday - Tuesday 07:30 - 19:00, Wednesday 08:00 - 18:00
- Participants who do not wish to be photographed have red lanyards; please note and respect
🙏 to
the Steph, Mouna and Garrett
colleagues, friends and learners at Schneider Electric, University of York and RForwards!
Posit team and especially Mine Çetinkaya-Rundel
- Ian!
- Experience 🍱 🥗 🌮 🍴 🕐
Prerequisites
You need a laptop with the following installed:
- R version ≥4.2 and RStudio ≥2023.03
- The following packages: {tidyverse}, {palmerpenguins}, {devtools}, {here}
🎬 Detailed instructions for installing these were covered in Prerequisites
Schedule
| Time | Activity |
|---|---|
| 09:00 - 10:30 | Functions 1 Introduction, vector and dataframe functions, embracing |
| 10:30 - 11:00 | ☕ Coffee break |
| 11:00 - 12:30 | Functions 2 Plot functions, style and side effects |
| 12:30 - 13:30 | 🍱 🥗 🌮 🍴 Lunch break |
| 13:30 - 15:00 | Iteration 1 Introduction and modifying multiple columns |
| 15:00 - 15:30 | ☕ Coffee break |
| 15:30 - 17:00 | Iteration 2 Reading and writing multiple files |
How we will work
-
stickies
🟦 I’m all good, I’m done
🟪 I could do with some help
Discord
no stupid questions
🎬 Action!
Learning Objectives
At the end of this section you will be able to:
- explain the rationale for writing functions
- write vector functions
- that take one or more vectors as input and output a vector
- that take one or more vectors as input and output a single value
- specify defaults for function argument
- write functions that take dataframes as input and output a dataframe
- using embracing to allow data masking and tidy selection within functions
Set up
Project
https://github.com/posit-conf-2023/programming-r-exercises
🎬 Create a Project:
> usethis::use_course("posit-conf-2023/programming-r-exercises")
✔ Downloading from 'https://github.com/posit-conf-2023/programming-r-exercises/zipball/HEAD'
Downloaded: 0.26 MB
✔ Download stored in 'C:/Users/er13/OneDrive - University of York/Desktop/Desktop/posit-conf-2023-programming-r-exercises-978baff.zip'
✔ Unpacking ZIP file into 'posit-conf-2023-programming-r-exercises-978baff/' (45 files extracted)
Shall we delete the ZIP file ('posit-conf-2023-programming-r-exercises-978baff.zip')?
1: Not now
2: Yeah
3: Nope🎬 Choose the option that means yes!
✔ Deleting 'posit-conf-2023-programming-r-exercises-978baff.zip'
✔ Opening project in RStudioRStudio will restart
Create a .R
usethis::use_r("functions-01")Packages
🎬 Load packages:
── Attaching core tidyverse packages ──────────────────────────────────────────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.2 ✔ readr 2.1.4
✔ forcats 1.0.0 ✔ stringr 1.5.0
✔ ggplot2 3.4.2 ✔ tibble 3.2.1
✔ lubridate 1.9.2 ✔ tidyr 1.3.0
✔ purrr 1.0.1 ── Conflicts ────────────────────────────────────────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package to force all conflicts to become errors'Load penguins
🎬 Load penguins data set
Rows: 344
Columns: 8
$ species <fct> Adelie, Adelie, Adelie, Adelie, Adelie, Adelie, Adel…
$ island <fct> Torgersen, Torgersen, Torgersen, Torgersen, Torgerse…
$ bill_length_mm <dbl> 39.1, 39.5, 40.3, NA, 36.7, 39.3, 38.9, 39.2, 34.1, …
$ bill_depth_mm <dbl> 18.7, 17.4, 18.0, NA, 19.3, 20.6, 17.8, 19.6, 18.1, …
$ flipper_length_mm <int> 181, 186, 195, NA, 193, 190, 181, 195, 193, 190, 186…
$ body_mass_g <int> 3750, 3800, 3250, NA, 3450, 3650, 3625, 4675, 3475, …
$ sex <fct> male, female, female, NA, female, male, female, male…
$ year <int> 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007…Why write functions?
Rationale
- impact from code: reach and clarity
- efficiency: update code in one place, decrease error rate, improve ability to reuse
Example
We have several measurements:
bill_length_mmbill_depth_mmflipper_length_mmbody_mass_g
These are on very different scales
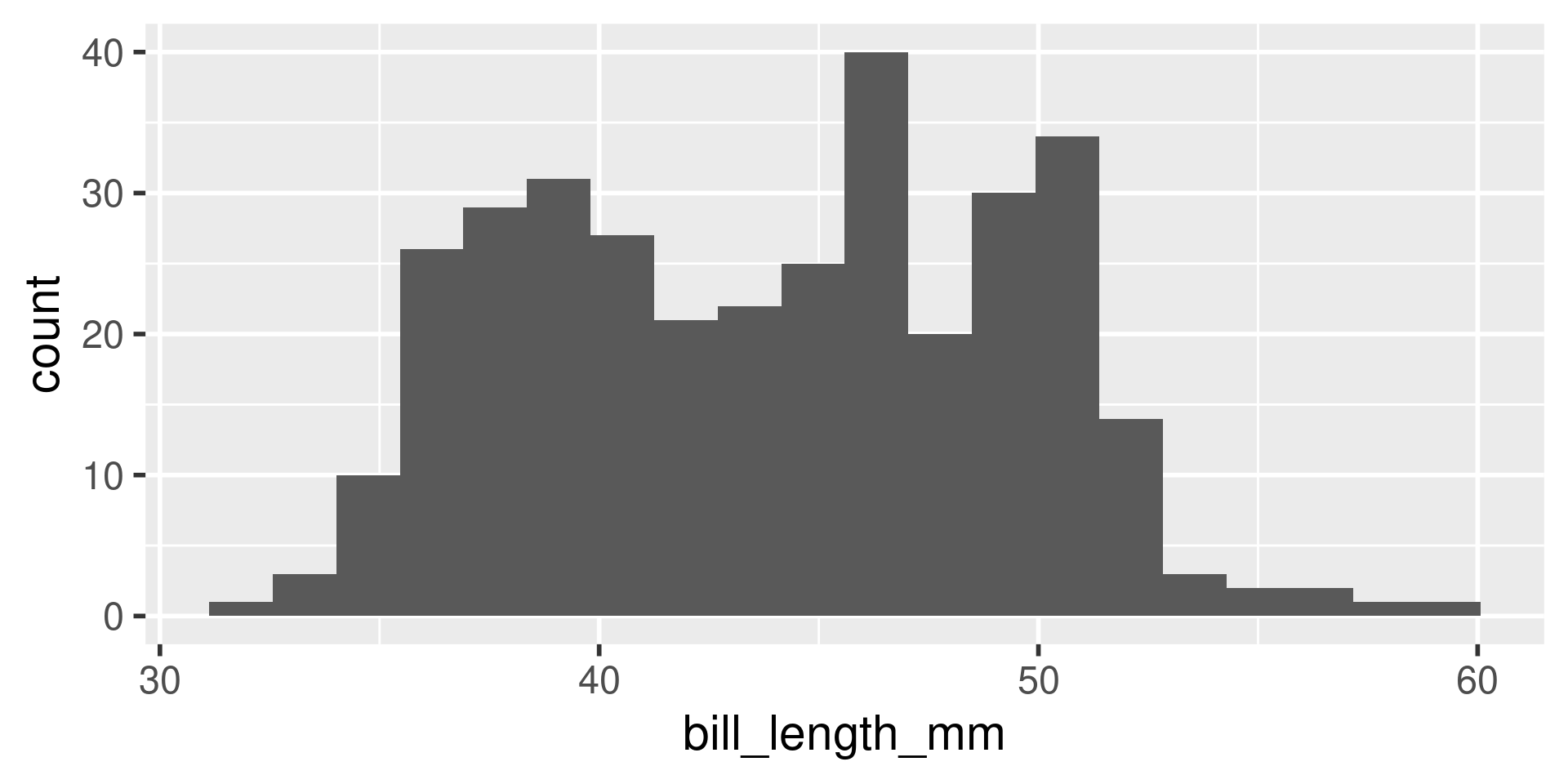
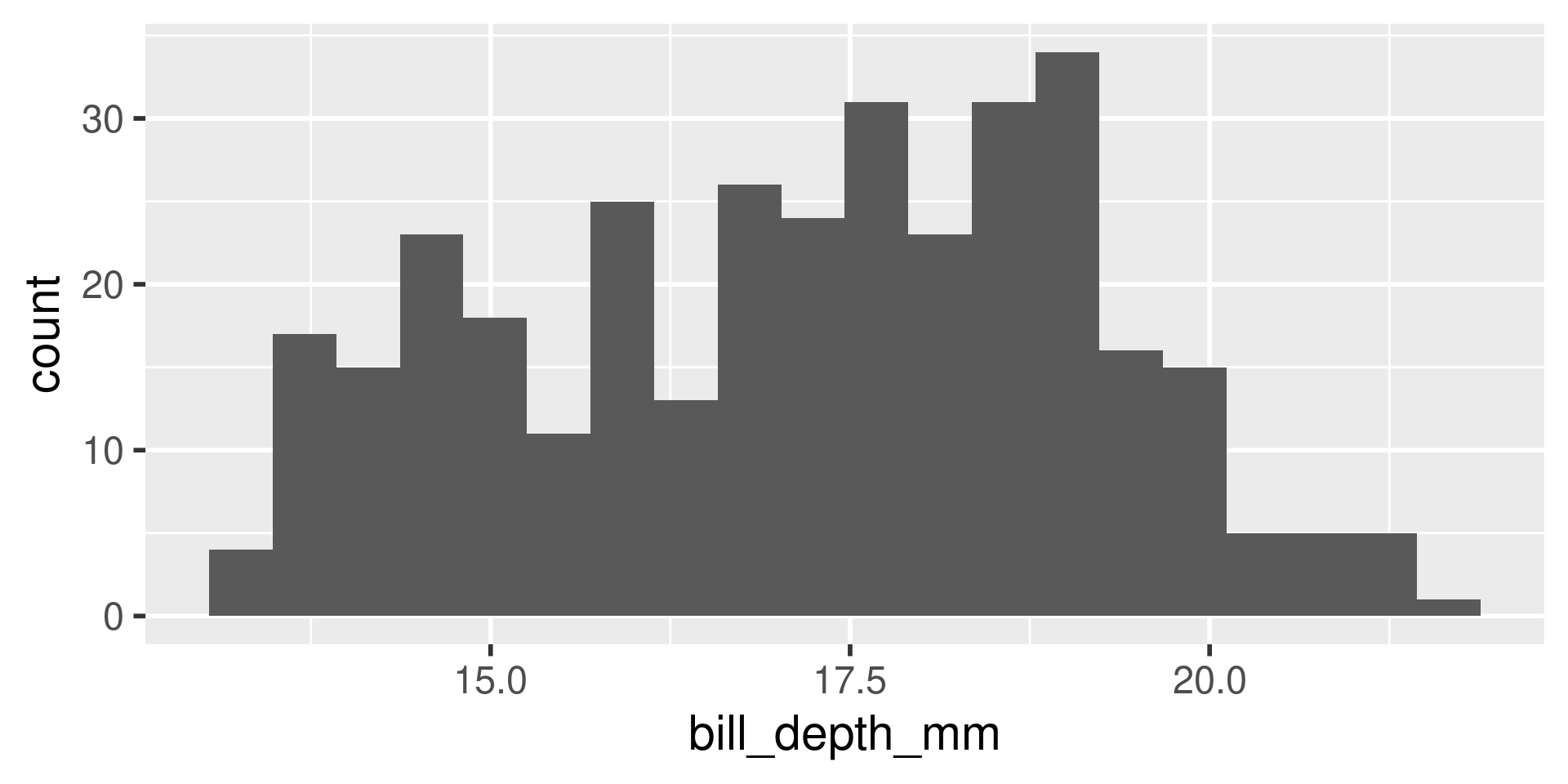
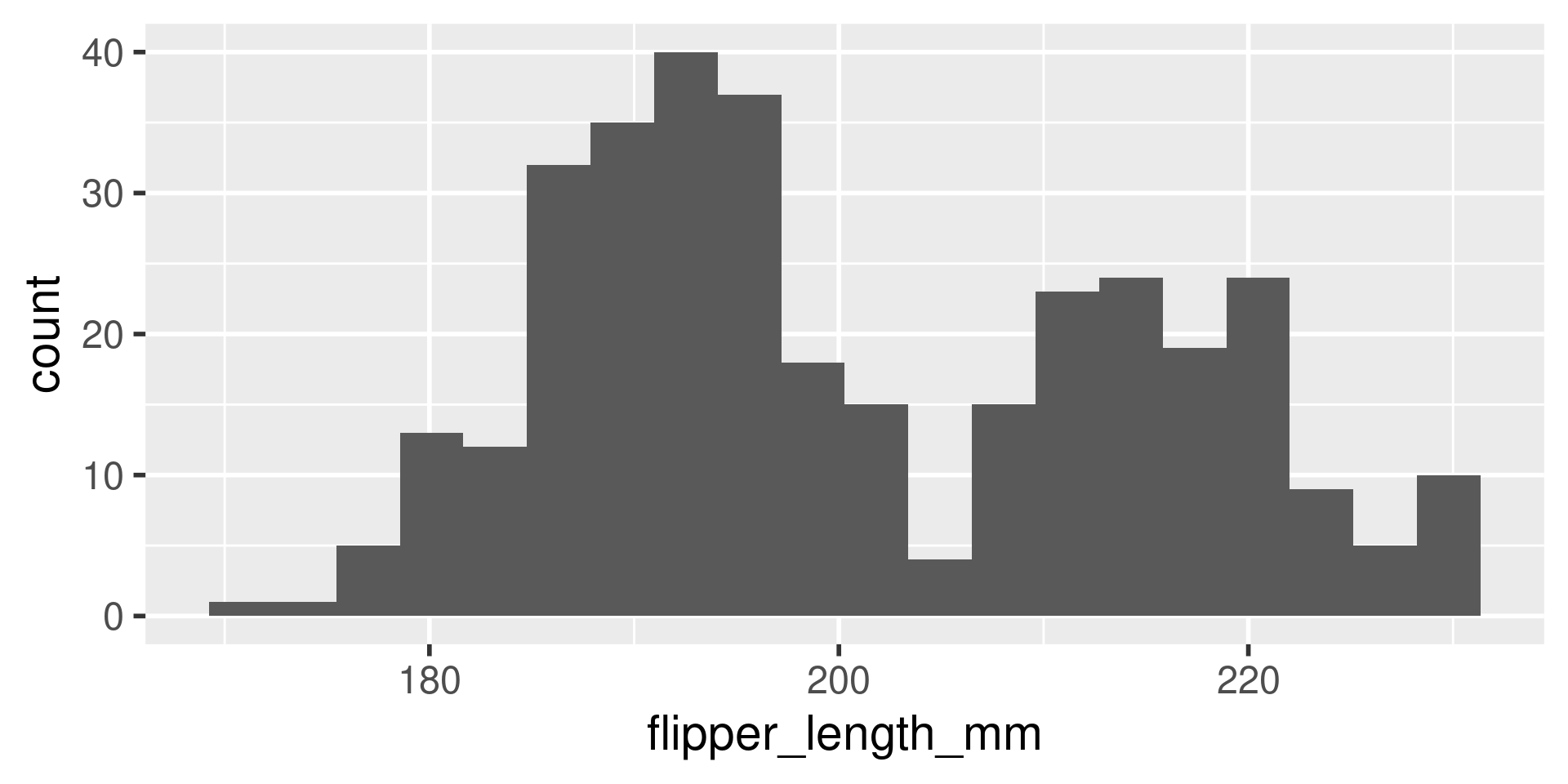
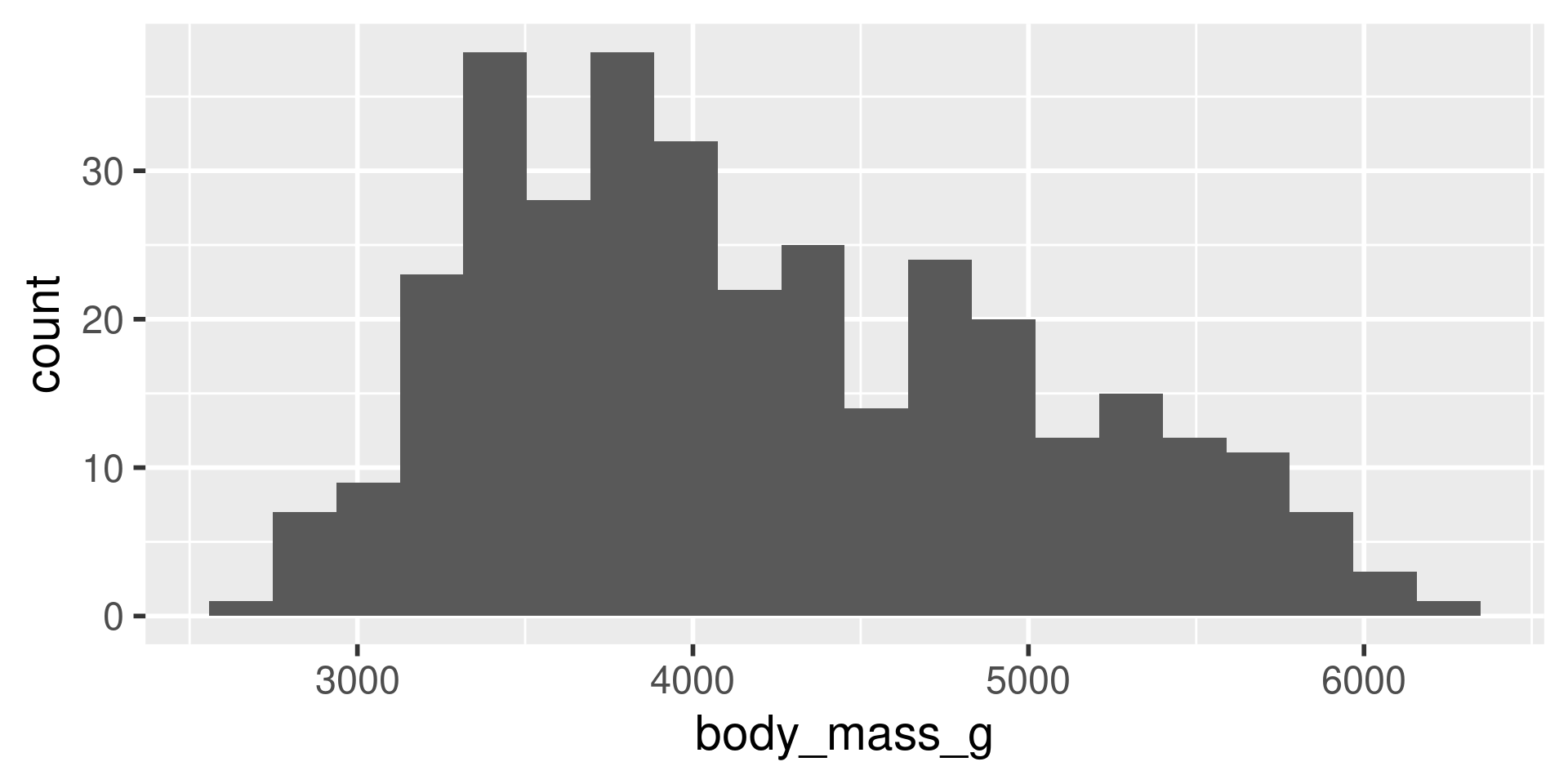
Example
difficult to plot on same axis or determine what value is large for that variable
A common solution is to apply a \(z\) score transformation to each variable.
Normalises the values to have a mean of 0 and a standard deviation of 1
\[z = \frac{x - \bar{x}}{s.d.}\]
Apply transformation
We can apply the same transformation to each variable:
penguins <- penguins |>
mutate(
z_bill_length_mm = (bill_length_mm - mean(bill_length_mm, na.rm = TRUE)) / sd(bill_length_mm, na.rm = TRUE),
z_bill_depth_mm = (bill_depth_mm - mean(bill_depth_mm, na.rm = TRUE)) / sd(bill_depth_mm, na.rm = TRUE),
z_flipper_length_mm = (flipper_length_mm - mean(flipper_length_mm, na.rm = TRUE)) / sd(flipper_length_mm, na.rm = TRUE),
z_body_mass_g = (body_mass_g - mean(body_mass_g, na.rm = TRUE)) / sd(body_mass_g, na.rm = TRUE)
)Long, unclear
(bill_length_mm - mean(bill_length_mm, na.rm = TRUE)) / sd(bill_length_mm, na.rm = TRUE)
- Quite a lot of code
- Difficult to determine what the transformation is
How to shorten and make more clear?
Coping and pasting
- is error prone
How to make fewer mistakes?
Writing a function:
- can be named to make transformation transparent
- will make code shorter
- can be reused
🔑️ You may think you have to write complex functions - you don’t! Start with the simple things.
Types of function
Types of function
We will cover two types of function
- vector functions: one of more vectors as input, one vector as output
- data frame functions: df as input and df as output
Types of function
We will cover two types of function
-
vector functions: one of more vectors as input, one vector as output
data frame functions: df as input and df as output
Vector functions
Output same length as input
- output same length as input
- work well in
mutate() - appropriate for the z-transformation example
General
To turn your code into a function you need:
- a name
- the arguments - which represent the bits that vary
- the code body for the function
Function name
Use a verb - The tidyverse style guide (Wickham, n.d.) but good advice regardless
Difficulty in naming? Should this be two or three functions?
What should we call the function we write to do a \(z\) score transformation?
Arguments
the input vector
additional arguments
Naming conventions
- x for the vector input
Example
\[z = \frac{x - \bar{x}}{s.d.}\]
penguins <- penguins |>
mutate(
z_bill_length_mm = (bill_length_mm - mean(bill_length_mm, na.rm = TRUE)) / sd(bill_length_mm, na.rm = TRUE),
z_bill_depth_mm = (bill_depth_mm - mean(bill_depth_mm, na.rm = TRUE)) / sd(bill_depth_mm, na.rm = TRUE),
z_flipper_length_mm = (flipper_length_mm - mean(flipper_length_mm, na.rm = TRUE)) / sd(flipper_length_mm, na.rm = TRUE),
z_body_mass_g = (body_mass_g - mean(body_mass_g, na.rm = TRUE)) / sd(body_mass_g, na.rm = TRUE)
)Example
Identify the arguments: the things that vary across calls
(bill_length_mm - mean(bill_length_mm, na.rm = TRUE)) / sd(bill_length_mm, na.rm = TRUE)
(bill_depth_mm - mean(bill_depth_mm, na.rm = TRUE)) / sd(bill_depth_mm, na.rm = TRUE)
(flipper_length_mm - mean(flipper_length_mm, na.rm = TRUE)) / sd(flipper_length_mm, na.rm = TRUE)
(body_mass_g - mean(body_mass_g, na.rm = TRUE)) / sd(body_mass_g, na.rm = TRUE)Example
Put into the template
Apply
Rewrite the call to mutate() as:
penguins <- penguins |>
mutate(
z_bill_length_mm = to_z(bill_length_mm),
z_bill_depth_mm = to_z(bill_depth_mm),
z_flipper_length_mm = to_z(flipper_length_mm),
z_body_mass_g = to_z(body_mass_g)
)Much shorter, much more clear.
A modification
mean() has a trim argument: mean(x, trim = 0, na.rm = FALSE, ...)
the fraction (0 to 0.5) of observations to be trimmed from each end of x before the mean is computed.
Suppose we want to specify the middle proportion left rather than the proportion trimmed from each end [^1]
A modification
A value of 0.1 for
trimtrims 0.1 from each end leaving 0.8 in the middletrim = (1 - middle)/2
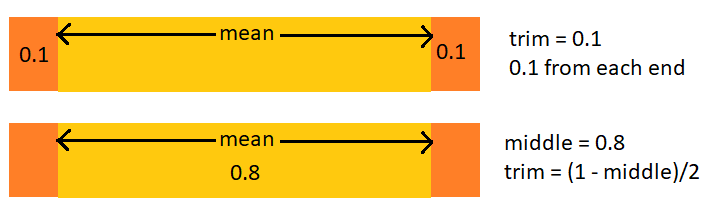
Trim is the proportion trimmed off each end; middle is what’s left
Add an argument
Try it out
to_z(penguins$bill_length_mm, middle = 0.2) [1] -0.92838057 -0.85511491 -0.70858359 NA -1.36797452 -0.89174774
[7] -0.96501340 -0.91006415 -1.84420131 -0.39720454 -1.16649396 -1.16649396
[13] -0.56205227 -1.01996264 -1.75261923 -1.38629094 -1.00164623 -0.30562246
[19] -1.78925206 0.33545205 -1.16649396 -1.18481038 -1.51450584 -1.09322830
[25] -0.98332981 -1.62440433 -0.65363435 -0.67195076 -1.14817755 -0.67195076
[31] -0.85511491 -1.27639245 -0.85511491 -0.59868510 -1.42292377 -0.91006415
[37] -0.98332981 -0.36057171 -1.20312679 -0.80016566 -1.40460735 -0.61700152
[43] -1.49618943 -0.01255983 -1.31302528 -0.83679849 -0.56205227 -1.22144320
[49] -1.49618943 -0.34225529 -0.83679849 -0.74521642 -1.67935358 -0.39720454
[55] -1.77093565 -0.50710303 -0.94669698 -0.65363435 -1.40460735 -1.20312679
[61] -1.55113867 -0.52541944 -1.20312679 -0.56205227 -1.42292377 -0.47047020
[67] -1.58777150 -0.56205227 -1.51450584 -0.43383737 -1.95409980 -0.81848208
[73] -0.83679849 0.29881922 -1.58777150 -0.25067322 -0.59868510 -1.27639245
[79] -1.45955660 -0.37888812 -1.75261923 -0.23235681 -1.36797452 -1.66103716
[85] -1.25807603 -0.52541944 -1.44124018 -1.33134169 -1.07491189 -0.96501340
[91] -1.55113867 -0.56205227 -1.86251772 -0.83679849 -1.45955660 -0.61700152
[97] -1.11154472 -0.70858359 -2.02736546 -0.17740756 -1.67935358 -0.58036869
[103] -1.18481038 -1.16649396 -1.14817755 -0.81848208 -1.01996264 -1.09322830
[109] -1.11154472 -0.17740756 -1.11154472 0.26218639 -0.81848208 -0.36057171
[115] -0.83679849 -0.26898963 -1.01996264 -1.25807603 -1.55113867 -0.56205227
[121] -1.45955660 -1.18481038 -0.72690000 -0.50710303 -1.64272075 -0.65363435
[127] -0.98332981 -0.48878661 -0.94669698 -0.01255983 -1.03827906 -0.19572398
[133] -1.34965811 -1.22144320 -1.11154472 -0.56205227 -1.56945509 -0.72690000
[139] -1.31302528 -0.81848208 -0.72690000 -0.65363435 -2.21052960 -0.63531793
[145] -1.25807603 -0.94669698 -0.91006415 -1.38629094 -1.49618943 -1.16649396
[151] -1.49618943 -0.48878661 0.35376847 1.06810865 0.82999525 1.06810865
[157] 0.62851469 0.42703413 0.22555357 0.46366696 -0.15909115 0.48198337
[163] -0.59868510 0.88494450 0.24386998 0.77504601 0.29881922 0.93989374
[169] -0.39720454 0.92157733 0.37208488 0.82999525 1.10474148 0.17060432
[175] 0.42703413 0.39040130 -0.23235681 0.35376847 0.06070583 0.66514752
[181] 0.73841318 1.06810865 0.57356545 -0.25067322 0.17060432 2.82648447
[187] 0.90326091 0.77504601 -0.28730605 0.04238942 -0.03087624 0.82999525
[193] -0.26898963 0.99484299 0.20723715 0.99484299 1.15969072 -0.10414190
[199] 0.24386998 1.15969072 0.13397149 0.18892074 0.44535054 0.79336242
[205] 0.17060432 1.08642506 0.42703413 0.15228791 -0.06750907 0.24386998
[211] -0.17740756 1.14137431 0.20723715 0.37208488 0.28050281 1.85571448
[217] 0.29881922 1.03147582 0.37208488 0.97652657 -0.12245832 1.19632355
[223] 0.64683111 0.40871771 0.73841318 0.42703413 0.40871771 0.81167884
[229] 0.61019828 1.26958921 0.18892074 0.18892074 0.90326091 1.52601902
[235] 0.59188186 1.06810865 0.13397149 1.21463997 -0.14077473 1.30622204
[241] 0.61019828 1.45275336 0.61019828 1.47106977 0.24386998 0.97652657
[247] 0.06070583 1.21463997 0.95821016 0.50029979 0.77504601 1.26958921
[253] 0.79336242 2.14877712 0.55524903 0.90326091 0.57356545 0.48198337
[259] -0.45215378 1.69086675 -0.15909115 0.72009677 1.15969072 1.03147582
[265] -0.12245832 1.34285487 0.37208488 2.00224580 0.06070583 0.84831167
[271] 0.55524903 NA 0.48198337 1.14137431 0.18892074 1.04979223
[277] 0.42703413 1.06810865 1.30622204 0.22555357 1.56265185 0.18892074
[283] 0.35376847 1.30622204 0.33545205 1.30622204 0.44535054 1.37948770
[289] 0.51861620 1.43443694 0.31713564 1.15969072 1.12305789 2.53342183
[295] 0.40871771 0.92157733 -0.32393888 0.79336242 -0.17740756 1.17800714
[301] 0.46366696 1.43443694 1.15969072 0.97652657 0.40871771 1.58096826
[307] -0.59868510 1.83739807 -0.30562246 1.25127279 1.01315940 0.61019828
[313] 0.62851469 1.43443694 0.50029979 1.70918316 0.88494450 0.37208488
[319] 1.23295638 0.24386998 1.23295638 1.21463997 1.08642506 0.88494450
[325] 1.34285487 1.03147582 0.72009677 1.32453845 0.28050281 1.19632355
[331] -0.30562246 1.47106977 0.18892074 0.93989374 1.10474148 0.26218639
[337] 1.41612053 0.48198337 0.28050281 2.13046071 -0.12245832 0.99484299
[343] 1.21463997 1.10474148But what if we forget?
to_z(penguins$bill_length_mm)Error in to_z(penguins$bill_length_mm): argument "middle" is missing, with no defaultGive a default
Give defaults whenever possible:
Try it out
to_z(penguins$bill_length_mm) [1] -0.88320467 -0.80993901 -0.66340769 NA -1.32279862 -0.84657184
[7] -0.91983750 -0.86488825 -1.79902541 -0.35202864 -1.12131806 -1.12131806
[13] -0.51687637 -0.97478674 -1.70744334 -1.34111504 -0.95647033 -0.26044656
[19] -1.74407616 0.38062795 -1.12131806 -1.13963448 -1.46932994 -1.04805240
[25] -0.93815391 -1.57922843 -0.60845845 -0.62677486 -1.10300165 -0.62677486
[31] -0.80993901 -1.23121655 -0.80993901 -0.55350920 -1.37774787 -0.86488825
[37] -0.93815391 -0.31539581 -1.15795089 -0.75498976 -1.35943145 -0.57182562
[43] -1.45101353 0.03261607 -1.26784938 -0.79162259 -0.51687637 -1.17626731
[49] -1.45101353 -0.29707939 -0.79162259 -0.70004052 -1.63417768 -0.35202864
[55] -1.72575975 -0.46192713 -0.90152108 -0.60845845 -1.35943145 -1.15795089
[61] -1.50596277 -0.48024354 -1.15795089 -0.51687637 -1.37774787 -0.42529430
[67] -1.54259560 -0.51687637 -1.46932994 -0.38866147 -1.90892390 -0.77330618
[73] -0.79162259 0.34399512 -1.54259560 -0.20549732 -0.55350920 -1.23121655
[79] -1.41438070 -0.33371222 -1.70744334 -0.18718091 -1.32279862 -1.61586126
[85] -1.21290014 -0.48024354 -1.39606428 -1.28616579 -1.02973599 -0.91983750
[91] -1.50596277 -0.51687637 -1.81734182 -0.79162259 -1.41438070 -0.57182562
[97] -1.06636882 -0.66340769 -1.98218956 -0.13223166 -1.63417768 -0.53519279
[103] -1.13963448 -1.12131806 -1.10300165 -0.77330618 -0.97478674 -1.04805240
[109] -1.06636882 -0.13223166 -1.06636882 0.30736229 -0.77330618 -0.31539581
[115] -0.79162259 -0.22381374 -0.97478674 -1.21290014 -1.50596277 -0.51687637
[121] -1.41438070 -1.13963448 -0.68172411 -0.46192713 -1.59754485 -0.60845845
[127] -0.93815391 -0.44361071 -0.90152108 0.03261607 -0.99310316 -0.15054808
[133] -1.30448221 -1.17626731 -1.06636882 -0.51687637 -1.52427919 -0.68172411
[139] -1.26784938 -0.77330618 -0.68172411 -0.60845845 -2.16535371 -0.59014203
[145] -1.21290014 -0.90152108 -0.86488825 -1.34111504 -1.45101353 -1.12131806
[151] -1.45101353 -0.44361071 0.39894437 1.11328455 0.87517115 1.11328455
[157] 0.67369059 0.47221003 0.27072946 0.50884286 -0.11391525 0.52715927
[163] -0.55350920 0.93012040 0.28904588 0.82022191 0.34399512 0.98506964
[169] -0.35202864 0.96675323 0.41726078 0.87517115 1.14991738 0.21578022
[175] 0.47221003 0.43557720 -0.18718091 0.39894437 0.10588173 0.71032342
[181] 0.78358908 1.11328455 0.61874135 -0.20549732 0.21578022 2.87166037
[187] 0.94843681 0.82022191 -0.24213015 0.08756532 0.01429966 0.87517115
[193] -0.22381374 1.04001889 0.25241305 1.04001889 1.20486662 -0.05896600
[199] 0.28904588 1.20486662 0.17914739 0.23409663 0.49052644 0.83853832
[205] 0.21578022 1.13160096 0.47221003 0.19746381 -0.02233317 0.28904588
[211] -0.13223166 1.18655021 0.25241305 0.41726078 0.32567871 1.90089038
[217] 0.34399512 1.07665172 0.41726078 1.02170247 -0.07728242 1.24149945
[223] 0.69200701 0.45389361 0.78358908 0.47221003 0.45389361 0.85685474
[229] 0.65537418 1.31476511 0.23409663 0.23409663 0.94843681 1.57119492
[235] 0.63705776 1.11328455 0.17914739 1.25981586 -0.09559883 1.35139794
[241] 0.65537418 1.49792926 0.65537418 1.51624567 0.28904588 1.02170247
[247] 0.10588173 1.25981586 1.00338606 0.54547569 0.82022191 1.31476511
[253] 0.83853832 2.19395302 0.60042493 0.94843681 0.61874135 0.52715927
[259] -0.40697788 1.73604265 -0.11391525 0.76527266 1.20486662 1.07665172
[265] -0.07728242 1.38803077 0.41726078 2.04742170 0.10588173 0.89348757
[271] 0.60042493 NA 0.52715927 1.18655021 0.23409663 1.09496813
[277] 0.47221003 1.11328455 1.35139794 0.27072946 1.60782775 0.23409663
[283] 0.39894437 1.35139794 0.38062795 1.35139794 0.49052644 1.42466360
[289] 0.56379210 1.47961284 0.36231154 1.20486662 1.16823379 2.57859773
[295] 0.45389361 0.96675323 -0.27876298 0.83853832 -0.13223166 1.22318303
[301] 0.50884286 1.47961284 1.20486662 1.02170247 0.45389361 1.62614416
[307] -0.55350920 1.88257397 -0.26044656 1.29644869 1.05833530 0.65537418
[313] 0.67369059 1.47961284 0.54547569 1.75435906 0.93012040 0.41726078
[319] 1.27813228 0.28904588 1.27813228 1.25981586 1.13160096 0.93012040
[325] 1.38803077 1.07665172 0.76527266 1.36971435 0.32567871 1.24149945
[331] -0.26044656 1.51624567 0.23409663 0.98506964 1.14991738 0.30736229
[337] 1.46129643 0.52715927 0.32567871 2.17563660 -0.07728242 1.04001889
[343] 1.25981586 1.14991738Your turn
🎬 Write a function that performs the Box-Cox power transformation using the value of (non-zero) lambda (\(\lambda\)) supplied.
Your turn: Box-Cox
\[bc = \frac{x^{\lambda} - 1}{\lambda} \text{ for }\lambda \ne 0\]
- Set the default \(\lambda = 1\)
- Still have time? Check \(\lambda \ne 0\)
- Still have time? Check and amend for:
\[ bc = \begin{cases} \frac{x^{\lambda} - 1}{\lambda} & \text{for }\lambda \ne 0\\ log(x) & \text{for }\lambda = 0 \end{cases} \]
A solution 1
to_box_cox <- function(x, lambda = 1) {
(x^lambda - 1) / lambda
}A solution 1 - test
A solution 1 - test
to_box_cox(vals, 0.3) |>
hist()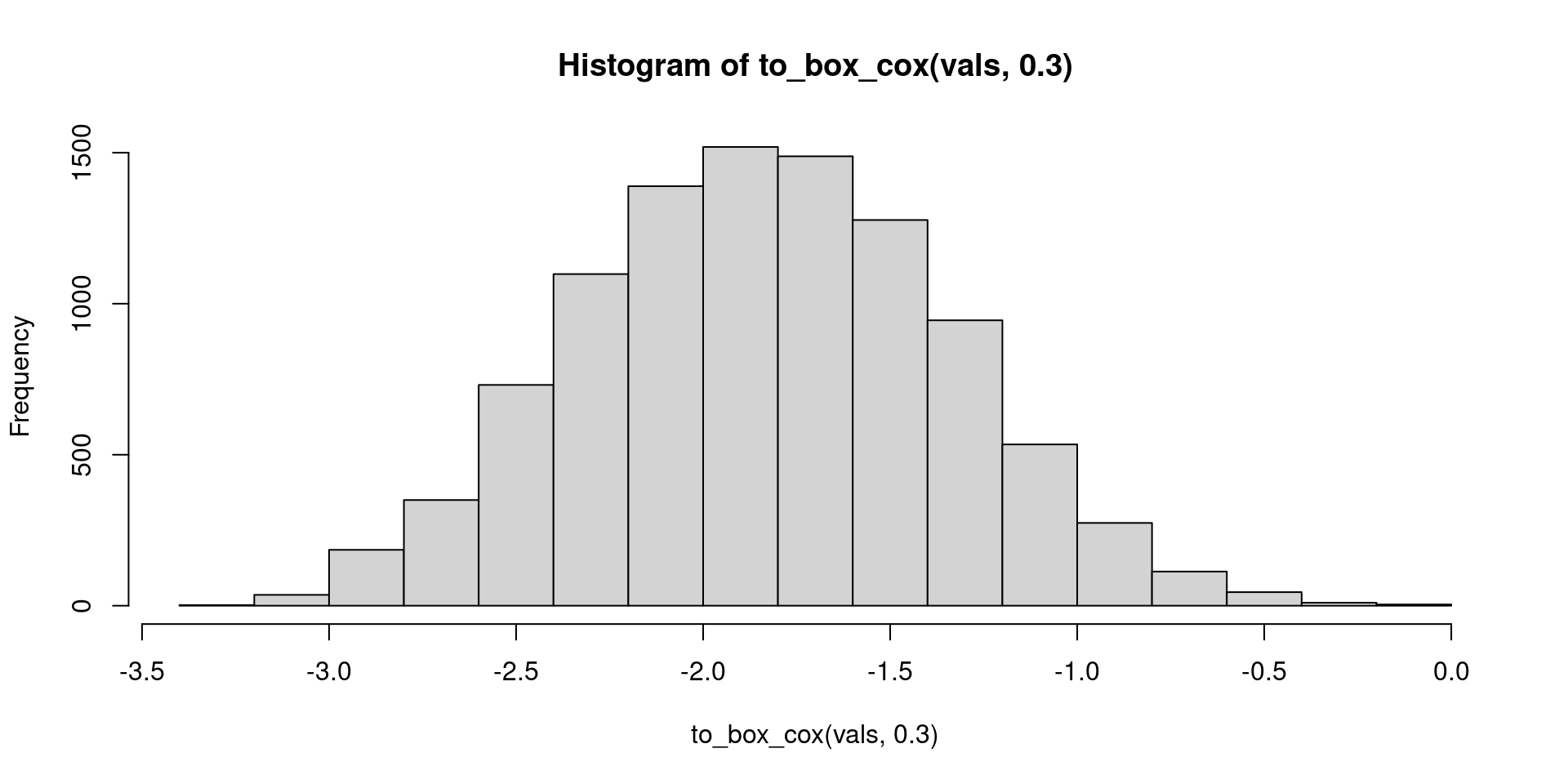
A solution 2
Check \(\lambda \ne 0\)
A solution 3
Check and amend for:
\[ bc = \begin{cases} \frac{x^{\lambda} - 1}{\lambda} & \text{for }\lambda \ne 0\\ log(x) & \text{for }\lambda = 0 \end{cases} \]
Types of function
We will cover two types of function
-
vector functions: one of more vectors as input, one vector as output
- ✔️ output same length as input.
ii. ➡️ summary functions: input is vector, output is a single value
data frame functions: df as input and df as output
Summary functions
- input is vector
- output is a single value
- could be used in
summarise()
Example
Write a function to compute the standard error of a sample.
\[s.e. = \frac{s.d.}{\sqrt{n}}\]
Example
Note: sum(TRUE) = 1 and sum(FALSE) = 0 Thus,sum(!is.na(x)) gives you the number of TRUE (i.e., the number of non-NA values) and is a bit shorter than length(x[!is.na(x)])
Try it out
🎬 Call the function on penguins$bill_length_mm
sd_error(penguins$bill_length_mm)[1] 0.2952205Or in a pipeline
penguins |>
summarise(se = sd_error(bill_length_mm))# A tibble: 1 × 1
se
<dbl>
1 0.295Your turn
🎬 Write a function to compute the sums of squares (sum of the squared deviations from the mean)
\[SS(x) = \sum{(x - \bar{x})^2}\]
or
\[SS(x) = s^2 * (n-1)\]
A solution - 1
🎬 Try it out
sum_sq(penguins$bill_length_mm)[1] 10164.21Types of function
We will cover two types of function
-
vector functions: one of more vectors as input, one vector as output
✔️ output same length as input.
✔️ summary functions: input is vector, output is a single value
2. ➡️ data frame functions: df as input and df as output
Dataframe functions
Dataframe functions
Dataframe as input and Dataframe as output
For example, we might summarise one of our columns like this:
penguins |>
summarise(mean = mean(bill_length_mm, na.rm = TRUE),
n = sum(!is.na(bill_length_mm)),
sd = sd(bill_length_mm, na.rm = TRUE),
se = sd_error(bill_length_mm))# A tibble: 1 × 4
mean n sd se
<dbl> <int> <dbl> <dbl>
1 43.9 342 5.46 0.295Output is a dataframe
Dataframe functions
and summarise several dataframes in the same way
Good candidate for a function to avoid repetitive code: my_summary()
Define my_summary() function
Use function
my_summary(penguins, bill_length_mm)Error in `summarise()`:
ℹ In argument: `mean = mean(column, na.rm = TRUE)`.
Caused by error:
! object 'bill_length_mm' not found😕
Tidy evaluation
tidyverse functions like dplyr::summarise() use “tidy evaluation” so you can refer to the names of variables inside dataframes. For example, you can use:
either
Or
rather than $ notation
This is known as data-masking: the dataframe environment masks the user environment by giving priority to the dataframe.
Data masking is great….
and makes life easier when working interactively
But not so useful in functions
Because of data-masking, summarise() in my_summary() is looking for a column literally called column in the dataframe that has been passed in. It is not looking in the variable column for the name of column you want to give it.
Fix my_summary() function
The solution is to use embracing: {{ var }}
- look inside
columnvariable - style with spaces
-
.groups = "drop"to avoid message and leave the data in an ungrouped state
Use function
my_summary(penguins, bill_length_mm)# A tibble: 1 × 4
mean n sd se
<dbl> <int> <dbl> <dbl>
1 43.9 342 5.46 0.295🎉
When to embrace?
When tidy evaluation is used
Your turn
🎬 Write a function to calculate the median, maximum and minimum values of a variable grouped by another variable.
A solution - 1
Your turn
🎬 Try it out
my_summary(penguins, bill_length_mm, species)# A tibble: 3 × 4
species median minimum maximum
<fct> <dbl> <dbl> <dbl>
1 Adelie 38.8 32.1 46
2 Chinstrap 49.6 40.9 58
3 Gentoo 47.3 40.9 59.6A solution - 2
Improvement: Have a default of NULL for the grouping variable
Your turn
🎬 Try it out
my_summary(penguins, bill_length_mm)# A tibble: 1 × 3
median minimum maximum
<dbl> <dbl> <dbl>
1 44.4 32.1 59.6Your turn
🎬 Try it out with more than one group
my_summary(penguins, bill_length_mm, c(species, island),)Error in my_summary(penguins, bill_length_mm, c(species, island), ): unused argument (alist())😕
A solution - 3
Use pick() which allows you to select a subset of columns inside a data masking function:
🎬 Try it out with more than one group
my_summary(penguins, bill_length_mm, c(species, island))# A tibble: 5 × 5
species island median minimum maximum
<fct> <fct> <dbl> <dbl> <dbl>
1 Adelie Biscoe 38.7 34.5 45.6
2 Adelie Dream 38.6 32.1 44.1
3 Adelie Torgersen 38.9 33.5 46
4 Chinstrap Dream 49.6 40.9 58
5 Gentoo Biscoe 47.3 40.9 59.6Extras
-
Short cuts:
- put cursor on a function call and press F2 to find its definition
- Ctrl+. opens section/file search
Summary ☕
Writing functions can make you more efficient and make your code more readable. This can be just for your benefit.
Vector functions take one of more vectors as input; output can be a vector (useful in
mutate()andfilter()) or a single value (useful insummarise())Dataframe functions take a dataframe as input and output a dataframe
Give arguments a default where possible
We use
{{ var }}embracing to manage data maskingWe use
pick()to select more than one variable